Biospecimen Core
Overview
PI: Bharat Thyagarajan
The LLFS Biospecimen Core has collected and stored a wide variety of biospecimens on LLFS participants from Visit 1 and Visit 2. At both visits, the LLFS biorepository consists of serum, plasma, PAXgene tudes (for evaluating RNA based biomarkers) and cryopreserved peripheral blood nuclear cells (PBMCs). The Advanced Research and Diagnostics Laboratory (ARDL) at the University of Minnesota has served as the centrl laboratory and biorepository for LLFS since the start of the study. In addition to maintaining the central biorepository, ARDL has also measured >40 protein and lipid based biomarkersin LLFS at baseline and subsets of these biomarkers have been measured at Visit 2. ARDL has successfully collaborated with the Centers for Inherited Disease (CIDR), Broad Institute, and the Washington University Genome Center to facilitate genotyping and sequencing efforts in LLFS using DNA and RNA samples from the LLFS biorepository.
The Biospecimen Core will build on its experience working with the LLFS Field Centers and the DMCC to coordinate biospecimen collection and processing during the third exam, develop and implement quality control (QC) measures to harmonize analyte measurements across multiple visits, harmonize biomarker measurement with other studies such as the Framingham Heart Study (FHS), the Health and Retirement Study (HRS), the New England Centenarian Study (NECS) and the Baltimore Longitudinal Study of Aging (BLSA) and coordinate measurement of multiple omics platforms using the extensive biorepository available in LLFS.
The specific aims of the Biospecimen Core are as follows:
Aim 1
Collect high quality biospecimens from (a) surviving LLFS participants attending a third in-person visit and (b) participants from the grandchild generation at Visit 3 (the initial visit for this generation).
Subaim 1.1: Develop a QC program for biospecimen collection.
Subaim 1.2: Develop a QC program for anallyte measurement.
Aim 2
Longitudinal measurement of aging and Alzheimer’s disease biomarkers.
Subaim 2.1: Measurement of aging and Alzheimer’s disease biomarkers.
Subaim 2.2: Identificaiton of novel protein based aging biomarkers.
Aim 3
Implement and harmonize multiple omics measurements in LLFS.
Subaim 3.1: Develop a QC program for external harmonization of omics data.
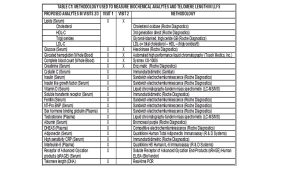
Known Biomarkers
Responsible for overseeing biospecimen collection, processing, and storage of specimens. Collection methods same for all visits. Participants are asked to be fasting. If no blood sample is obtained, ask participants for saliva sample (Oragene Inc saliva collection kit). The quality control involves monitoring the completeness of blood collection at all sites, as well as the receipt of samples to the Central Lab (ARDL in MN) within 48 hours of collection. Blind duplicates are also included as part of the QC program as a way to estimate reproducibility and reduce/correct any potential batch effects of our bioassays. Additionally responsible for harmonization across the three LLFS visit as well as external studies.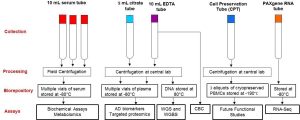
Genomic Group
Led by Dr. Nathan Stitziel of Washington University. Oversees the whole genome sequencing pipeline run out of the McDonnell Genome Institute (MGI) at Washington University. Includes the sequencing protocols and quality control. Guides the DMCC in creating clean WGS data for release for analyses. Emulates the QC protocol of TopMED as closely as possible, with some additional steps due to the familial natures of LLFS data. Analyses will also incorporate the insight of Dr. Stitziel’s lab through the extensive use of working group meetings.
Metabolome Group
Led by Dr. Gary Patti of Washington University. The Patti lab will run untargeted mass spectrometry on all LLFS participant samples at all longitudinal time points. They will also oversee the quality control and provide an analysis dataset for distribution to the Data Management and Coordinating Center. Analyses will also incorporate the insight of the Patti lab through the extensive use of working group meetings.
Methylome Group
Led by Dr. Ting Wang of Washington University. Dr. Wang’s lab will oversee the methods and quality control for whole genome bisulfite sequencing of LLFS participants which will be completed at the McDonnell Genome Institute (MGI). Sequencing and quality control protocols will be decided upon in collaboration with Dr. Wang. Analyses will also incorporate insight from Dr. Wang’s laboratory, through the extensive use of working group meetings.
Proteome Group
Led by Dr. Andrew Emili of Boston University. The Boston University group has prior expertise in the generation and analysis of proteomic data. Together in collaboration with the LLFS Central Blood Laboratory in Minnesota, they will create the proteomic pipelines for data generation and quality control. Analyses will also incorporate their insight through the extensive use of working group meetings.
Transcriptome Group
Led by Dr. Michael Brent of Washington University. The RNA sequence will be obtained in collaboration with the McDonnell Genome Institute, the LLFS Central Blood Laboratory in Minnesota, and Dr. Michael Brent. All protocols and procedures are being created for data generation. Quality control will be under the direction of Dr. Michael Brent, and analyses will also incorporate the Brent’s lab insight into the data through extensive use of working group meetings.