What are the secrets to a long and healthy Life?
Some families have the good fortune to live very
long lives . . .We want to know about those families!
Welcome to Long Life Family Study
Recent Press Releases Regarding LLFS and PacBio:
Additional Coverage
Globe Newswire Press Release Coverage
BioSpace Press Release Coverage
AInvest Press Release Coverage
GenomeWeb Press Release Coverage
Now recruiting new families for Visit 4!!!
Calculate your family’s exceptional longevity score here to see how exceptional your family may be!
If your family’s score is greater than 7 our staff will reach out for further information.

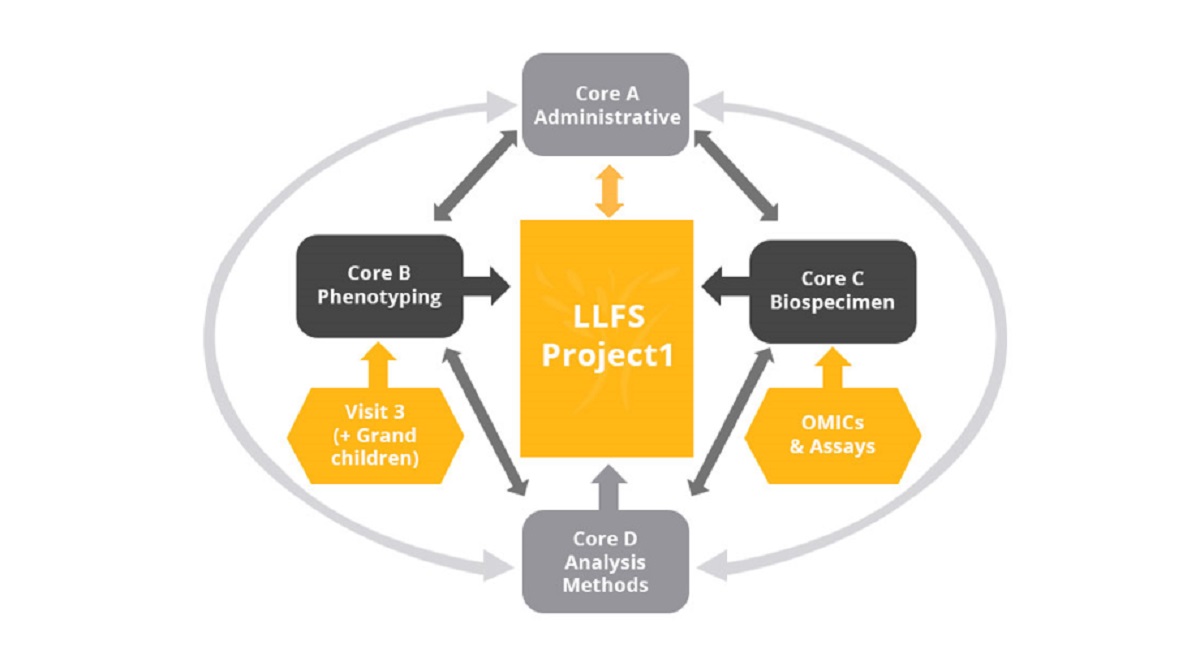
The multicenter LLFS originally enrolled and studied a unique sample of 4,953 participants in 539 pedigrees in the USA and Denmark which are enriched for familial EL. Through three extensive in-home visits, these families possess key healthy aging phenotypes (HAPs) in major domains of the aging process (cognitive, cardiovascular, metabolic, inflammatory, etc.). Over the course of the three visits, LLFS has now enrolled 5,438 individuals from the original 539 pedigrees, including grandchildren, making LLFS a three-generational study with longitudinal data spanning close to 20 years.
The measured phenotypes are highly heritable cross-sectionally and longitudinally; however considerable familial phenotypic heterogeneity is present. To better understand this heterogeneity and the why and how these families are protected we: continue annual follow-up with existing participants (telephone, mail, or online); combine linkage and association analyses to identify rare and lineage specific variants for cross-sectional and longitudinal HAPs and EL and their interaction with lifestyle exposures; perform comprehensive OMICs on LLFS pedigrees to discover biologic mechanisms leading to the heterogeneous familial patterns of HAPs and EL in LLFS pedigrees, and discover additional causal variants. Finally, we will combine all the data using systems biology and data integration to more comprehensively explain the biology of healthy aging.
The purpose of the LLFS is to study families in the United States and Denmark that are ALL enriched for exceptional longevity (EL). EL is a complex trait that is likely influenced by multiple genes with small effects interacting with lifetime exposures. The knowledge gained from studying these families will be in why and how they are protected and thus living exceptionally healthy long lives.
This project is supported by the National Institutes of Health’s National Institute on Aging, grant U19AG063893 and 2U19AG063893-06.
Alzheimer’s Disease Related Research Findings from LLFS
Xicota, Barral, and colleagues (https://pubmed.ncbi.nlm.nih.gov/38380866/) performed a genome wide search using the short read whole genome sequence (WGS) data from the LLFS dataset (n=3475) to understand the genetic contributions to delayed onset of late onset AD (LOAD) and confirmed the association in 6 external datasets (n=14,260). We identified several tightly linked SNPs in the MTUS2 gene (rs73154407, p = 7.6 × 10−9) that were significantly associated with LOAD, and the association of MTUS2 variants with LOAD was confirmed in 5 independent datasets. Subsequently, this study showed that the allelic association was stronger in those with high levels of Aβ42/40 ratio than in those with lower levels of Aβ42/40 ratio. This finding sheds light on AD biology in that MTUS2 encodes a microtubule associated protein implicated in the development and function of the nervous system, making it a plausible candidate to investigate LOAD biology.
Genes associated with physiological dysregulation may play a role in AD onset. Arbeev and colleagues (https://pubmed.ncbi.nlm.nih.gov/32235003/) found that genes associated with the age-related increase in physiological dysregulation (PD) frequently represented pathways implicated in axon guidance and synaptic function, which in turn were linked to AD and related traits (e.g., amyloid, tau, neurodegeneration). As a follow-up, Arbeev et al (https://pubmed.ncbi.nlm.nih.gov/37719713/) then investigated the hypothesis that genes involved in PD and axon guidance/synapse function may jointly influence onset of AD. We found significant interactions between SNPs in the UNC5C and CNTN6, and PLXNA4 and EPHB2 genes that influenced AD onset in both datasets. Our findings suggest the joint contribution of genes involved in PD and axon guidance/synapse function (essential for the maintenance of complex neural networks) to AD development.
Welcome to Long Life Family Study
Key Publications
NIA news and tools
- The latest research news from the National Institute on Aging at NIH: https://www.nia.nih.gov/news/news-releases
- NIA Events link: https://www.nia.nih.gov/news/events
- Lifelong exercise promotes brain health in older adults: https://www.nia.nih.gov/news/lifelong-exercise-promotes-brain-health-older-adults
- Maintaining mobility and preventing disability are key to living independently as we age: https://www.nia.nih.gov/news/maintaining-mobility-and-preventing-disability-are-key-living-independently-we-age
- Better executive function predicts less mobility decline after a fall: https://www.nia.nih.gov/news/better-executive-function-predicts-less-mobility-decline-after-fall
- Higher daily step count linked with lower all-cause mortality: https://www.nia.nih.gov/news/higher-daily-step-count-linked-lower-all-cause-mortality
- NIA Featured Research: https://www.nia.nih.gov/news/research-highlights
Publications
- Whole genome-wide sequence analysis of long-lived families (long Life Family Study) identifies MTUS2 gene associated with late-onset Alzheimer’s disease: https://pubmed.ncbi.nlm.nih.gov/38380866/
- Mechanisms underlying familial aggregation of exceptional health and survival: A three-generation cohort study: https://pubmed.ncbi.nlm.nih.gov/32886847/
- Patterns of multi-domain cognitive aging in participants of LLFS: https://pubmed.ncbi.nlm.nih.gov/32514870/
- Gene Discovery for high-density lipoprotein cholesterol level change over time in prospective family studies: https://pubmed.ncbi.nlm.nih.gov/32109663/
- Genome-wide linkage analysis of carotid artery traits in exceptionally long-lived families: https://pubmed.ncbi.nlm.nih.gov/31634740/
- More LLFS publications: link to LLFS publications page on website
Related Links
Tools
- National Institute on Aging (NIA): https://www.nia.nih.gov/
- Longevity Genomics: https://www.longevitygenomics.org/
- Longevity GWAS Catalog: https://www.ebi.ac.uk/gwas/efotraits/EFO_0004300